Scientists Unveiled and Characterized Four Episodes of Large-Scale Gene Duplications in the Evolutionary Past of Mosses
2020-11-05
Gene duplications provided the genetic raw materials for evolutionary innovation and are considered important driving forces in diversification and evolution. The evolutionary past of land plants is characterized by recurrent ancestral paleopolyploidy events (i.e. ancestral Whole Genome Duplications). The duplicated genes in both the model plant Arabidopsis and rice can be traced back to five rounds of polyploidy. Bryophytes (including mosses), which branched away from other land plants almost 500 million years ago, represented a key group occupying an important phylogenetic position in land plant (embryophyte) evolution. However, such important events in the evolutionary past of mosses has not been investigated.
To resolve an accurate phylogeny of mosses and the phylogenetic positioning of ancestral genomic duplication events, an international team was formed by researchers from the Chinese Academy of Sciences, Hong Kong Baptist University, The Chinese University of Hong Kong and University of Missouri. Their results are now published in Journal of Systematics and Evolution.
Phylotranscriptomic analyses of nearly 30 moss transcriptomes was utilized to probe single-copy gene families and generate reliable species phylogenies. This was coupled with a large-scale phylogenomic analysis investigating duplication signals from more than five thousand of gene trees to determine the phylogenetic positioning of ancestral genomic duplication events.
As a result, two branches with large numbers of gene duplications were elucidated by phylogenomic analyses, one in the ancestry of all mosses and another before the separation of the Bryopsida, Polytrichopsida, and Tetraphidopsida. The analysis of the phylogenetic progression of duplicated paralogs retained on genomic syntenic regions in the Physcomitrella patens genome confirmed that the whole‐genome duplication events WGD1 and WGD2 were re‐recognized as the ψ event and the Funarioideae duplication event, respectively. The ψ polyploidy event was tightly associated with the early diversification of Bryopsida, in the ancestor of Bryidae, Dicranidae, Timmiidae, and Funariidae.
Together, four branches with large numbers of gene duplications were unveiled in the evolutionary past of P. patens. Gene retention patterns following the four large‐scale duplications in different moss lineages were analyzed and discussed. Recurrent significant retention of stress‐related genes may have contributed to their adaption to distinct ecological environments and the evolutionary success of this early diverging land plant lineage.
This study was jointly supported by the NSFC-Xinjiang Key Project and National Science Foundation. The State Key Laboratory of Agrobiotechnology and School of Life Sciences of Chinese University of Hong Kong provided the computational resources for this project.
The paper was published in Journal of Systematics and Evolution, entitled "Ancestral gene duplications in mosses characterized by integrated phylogenomic analyses".
Article link: https://onlinelibrary.wiley.com/doi/10.1111/jse.12683
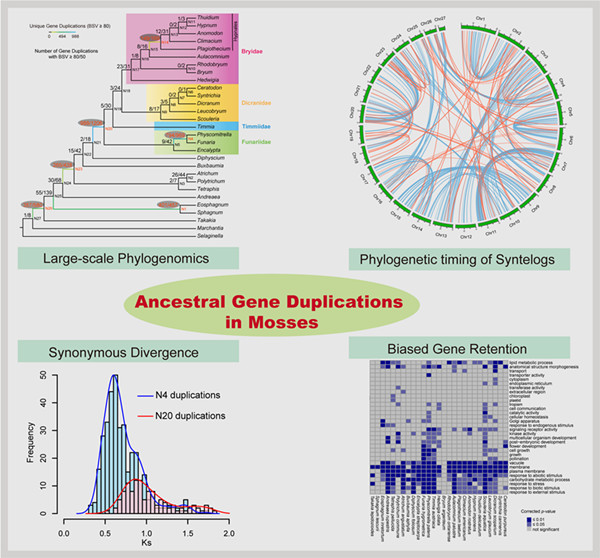
Figure: An integrated phylogenomic approach was employed to unveil and characterize the ancient gene duplications in mosses
Contact: LIU Jie, Xinjiang Institute of Ecology and Geography
E-mail: liujie@ms.xjb.ac.cn



